Translational Pharmacology
Branch of pharmacology that tries to bridge the gap between the basic molecular research studies to the clinical trials.
Translational Pharmacology, translational research and translational medicine are the interchangeable terminologies where
- the word translational focuses on development of a new drug which deals with the patient needs and targets to deal the specific issues.
translational research is an inter-disciplinary branch of a biomedical field that are greatly supported by three main pillars
- bench side (laboratory research),
- bed side (clinical practice) and
- the community (population needs).
Translational pharmacology includes:
- predictive experimental disease models;
- markers to assess the activity of drug candidates on targets;
- surrogate measures and biomarkers to detect disease initiation and progression; and
- sensitive and robust clinical measurements
There exists a gap between the need, development and the availability of the medicines.
specific systematic attempts reproduce preclinical findings, with very low success rates à failures are attributed to a variety of factors, including
- poorly specified methods,
- variable context-dependent behavior of tools and cell lines,
- inadequate experimental power,
several systematic analyses of the causes of major pharma pipeline attrition up to phase 2 of development.
- lack of efficacy as the cause of failure in approximately 50% of drug candidates.
Translational pharmacology helps to bridge this gap.
For academia, translational pharmacology encourages the individuals in the field of research, to test their novel ideas, that are generated from basic investigation hoping for their clinical applications.
While for physicians or the clinical practitioners it facilitates to capture the research benefits and to understand what is known and what is practiced.
While, as for the commercial pharmaceutical industry it measures assessment of the new entities at earlier phases, to identify the problems early, so as to solve them at an early stage to reduce the cost of product development.
The bench to bedside concept
Proper design, execution and reporting of animal model results help to make preclinical data more reproducible and translatable to the clinic. Hence, critical evaluation of the face and predictive validity of these models is important. Reversely, clinical bedside findings that were not predicted by animal testing should be back translated and used to refine the animal models.
Objectives
- to discover the origin, pathway and mechanism of the diseases including the responsible biomarkers, to discover and
- develop new diagnostic and therapeutic measures and to discover the newer drugs in short duration of time.
- The goal à not simply to design preclinical studies to demonstrate safety for first-in-human clinical administration, but to design studies that, together with Phase 1 clinical data, will be used to maximize the chances of success in the Phase 2 and Phase 3 clinical trials.
Translational Medicine department has three guiding principles:
(1) pick the right targets,
(2) develop the right biomarkers to measure target modulation, and
(3) test therapeutic hypotheses as safely, quickly and efficiently as possible.
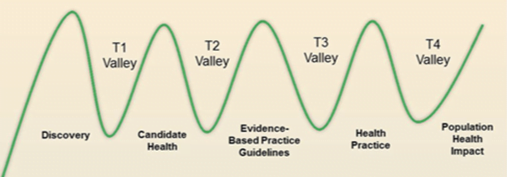
Phases of translational research
- Pharmacology research usually translates in vitro pharmacology experiments or animal studies to human proof of principle or phase I clinical studies. This is often referred as the T1 translational research.
- The clinical pharmacokinetic (PK) or pharmacodynamic (PD) studies usually translate and sometimes even guide the phases II and III clinical studies. This is called T2 translational research.
- The T3 translational research focuses on the practice, including comparative effectiveness research, postmarketing studies, and clinical outcome research,
- T4 translational research deals with population level outcome research, monitoring of morbidity, mortality, benefits, and risk, and impacts of policy and change.
- Usually, T3 and T4 fall into the pharmacoepidemiology research domain.
Valley of death
Translating the knowledge from biomedical science into clinical applications that help patients has been compared to crossing a valley of death because of the many issues that separate the bench from the bedside and threaten to stall progress
Next generation translational pharmacology
emergence of big data, increased accessibility of institutional electronic medical records, the availability of health insurance claims data through health information technology commercially, and many public in vitro drug screening data sources.
Unlike the conventional T1 to T4 translational research, these translational studies, driven by the big data, reversely connect the pharmaco-epidemiology evidences with in vitro pharmacology experiments.
Examples of application of translational pharmacology
- In vivo studies: Giving due importance to the time duration of treatment, animal characteristics, sample size and validity while planning an animal study is a good example of translational pharmacology.
- In vitro studies: Drug screening can be improved with the use of human cells of phenotype most relevant to the condition, ideally being derived from patients (representative of the disease stage being targeted), and then cultured in the most relevant conditions.
- In silico studies1: The in-silico models include developing and validating complex mathematical models that are capable of reflecting the human diseases and response to the therapeutic intervention. In- silico studies have the potential to speed the rate of discovery of a new molecule which reduces the need for expensive laboratory work and clinical trials for e.g. in 2010 using protein docking algorithm EADock, potential inhibitors to an enzyme associated cancer activity were found by in-silico studies.
- PK:PD Modelling8: Translational research would be helpful to prepare a model based on the pharmacokinetic and pharmacodynamic studies so that the safety and efficacy of newly introduced drug can be assessed prior to its use in the clinical trials. This can be achieved with the help of the mathematical tool called pharmacometrics, which is a branch of science that involves mathematical models of biology, pharmacology and disease. It also helps to describe the interaction between the xenobiotics and the patients which may include beneficial and harmful effects. PK:PD modelling approach was used to inform the dosing of the antiviral agent, Tamiflu, in pediatric application.
- Big data: Big data sources include observational (e.g. electronic medical/health records, the FDA adverse event reporting system etc.) and databases (e.g. DrugBank, PubMed, PharmGKB, LINCS). For example, the observational medical outcomes partnership (OMOP) recently attempted to integrate multiple observational databases through a common data model (CDM) by normalizing the dictionaries of medications, diagnoses and laboratory tests across all its (OMOP) databases.
- Adverse Drug Event (ADE) Reporting4: Although ADEs have been standardized through MedDRA (a medical terminology database for adverse drug events, www.meddra.org), whose utility in drug labelling has been well accepted, MedDRA still lacks sufficient annotation schemes to link an ADE with its molecular mechanism(s). Ontology deals with questions concerning what entities exist or can be said to exist, how such entities can be grouped or related within a hierarchy, and how they can be subdivided according to their similarities and differences. In pharmacology research, a properly constructed ontology will facilitate the proper presentation of drugs and ADE data, either from large clinical data- bases or from the literature. Duke et al. investigated drug interactions, using a local version of the OMOP database at Indiana University, to identify successfully six novel drug interaction pairs that significantly increased myopathy risk above a mere additive risk from two single drugs.
- Data mining9: The contributions of text mining to clinical pharmacology research arise mainly from two sources, medical records and research literature. Xu et al., developed Natural Language Processing (NLP) algorithms to extract medication data (i.e. drug names and dose information) from clinical notes, with significant high performance. Using this NLP, they successfully reproduced the pharmacogenetic effects of VKORC1 and CYP2C9 on warfarin weekly dosing.
- Omics10: Omics means profiling. The various subdisciplines under omics include transcriptomics (mRNA), proteomics (proteins), genomics and metabolomics (metabolites), amongst a host of other emerging areas. Proteomics are emerging biomarkers in preclinical and clinical research, a process in which the translation and back translation is crucial in order to improve the patient outcomes.
Examples à nutrigenomics, human microbiome.
